Ever wonder how scientists study our genetic code to help understand diseases, trace ancestry, and more? It is done by DNA Technology.
DNA analysis lies at the heart of major breakthroughs, but the techniques involved can sound intricate.
This post breaks down the standard DNA testing workflow in a quiz format while providing detailed explanations of each step.
By the end, you’ll not only be able to arrange the key terms correctly but also gain a deeper grasp of this transformative field.
Extraction: Isolating the Precious Genetic Material through DNA Technology
The first stage, DNA Extraction, aims to separate genes from cells in a pure form ready for manipulation.
At the core of every nucleated cell is a nucleus containing 46 tightly coiled chromosomes made of DNA wound around histone proteins.
Packaged this densely, the vital strands remain inaccessible.
Extraction methods therefore target cell lysis or controlled rupture of their outer phospholipid membrane and nuclear envelope.
Common disruption techniques include heat, chemical detergents, and enzymatic digestion. Heat lysis warms samples above DNA’s melting temperature to break hydrogen bonds stabilizing membrane structure.
Alternately, enzymes like lysozyme or Proteinase K are employed to degrade proteins making up the cell wall.
Detergent lysis works by solubilizing membrane lipids, dissolving them into micelles. Popular options include sodium dodecyl sulfate (SDS) and Triton X 100.
The addition of a suitable buffer maintains pH balance as these surfactants strip away membrane integrity.
After lysis, microcentrifugation separates cell debris from the extract solution suspended with released DNA plus RNA if not treated with RNase.
Repeated centrifugation and washing purifies the final extract for quantification and storage.
Lack of DNA extraction would render subsequent analysis impossible by leaving genetic material stuck inside cells, so this initial isolation is essential to the overall process.
A variety of kits now automate extraction for different sample types like blood, tissue, buccal swabs or microbiota in food/environmental samples.
PCR Amplification: Enabling Detection Down to a Single Molecule
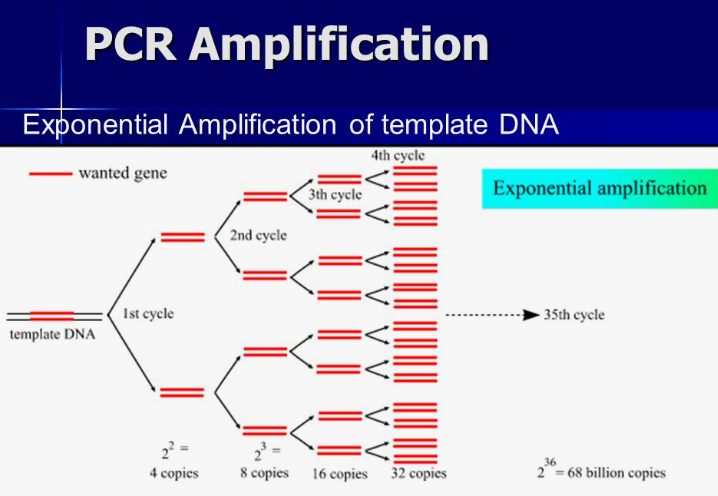
Once DNA is extracted from its cellular prison, the next major step multiplies it exponentially via Polymerase Chain Reaction, often called PCR.
Developed in 1983, this technique allows scientists to detect and analyze even minute starting quantities approaching a single molecule.
The secret lies in thermally cycling a reaction through temperature changes that induce structural changes in DNA.
When cooled below its melting point, double stranded DNA “zips up” due to hydrogen bonding favoring its helical conformation.
Raising temperature above the melting point “unzips” these bonds, physically separating the strands.
During PCR, three thermal cycles are repeated 20 to 40 times: denaturation at 96°C breaking base pairs, annealing primers around 55 to 65°C, then extension at 72°C using DNA polymerase to synthesize the complementary strand for each original template.
Special primers target delimited genomic regions for duplication within the linear amplifying phase.
Each cycle doubles the number of target DNA fragments, resulting in exponential growth up to billions of copies from picogram amounts in just a few hours.
This remarkable amplification boosts samples above detection thresholds otherwise too minute to study.
Without PCR, downstream techniques like sequencing and microarrays would lack the sensitivity afforded by such enrichment.
Furthermore, quantitative PCR variants reliably quantify initial DNA or RNA levels accurately down to a single molecule.
Sequencing: Reading the As, Ts, Cs and Gs
Amplified DNA undergoes sequencing to discern the precise order of its four nucleobases adenine, thymine, cytosine and guanine comprising the genetic alphabet.
Pioneering techniques like Sanger sequencing generate DNA fragments of defined lengths terminated by fluorescent dyes signaling each base.
More recent next generation platforms massively parallelized the process with synthesis or sequencing by ligation approaches.
Illumina sequencing by synthesis attaches thousands of single DNA strands to a flow cell where DNA polymerase with fluorescently labeled reversible terminator nucleotides assemble new strands in parallel.
Pacific Biosciences uses zero mode waveguides to monitor individual polymerase molecules as they incorporate fluorescently tagged bases in real time.
Oxford Nanopore sequencing takes a molecule sized protein pore embedded in a synthetic membrane and threads DNA through it like beads on a string.
As each base passes, it alters electrical signals uniquely identifying A, T, C or G. Such third generation techniques routinely sequence entire genomes in days with throughput exceeding hundreds of gigabases annually.
Data Analysis: Making Biological Sense of Raw Genetic Code
The final analysis phase makes biological meaning of raw sequence data comprising millions or billions of overlapping DNA snippets.
Assembly algorithms first reconstruct full chromosomes or organelle sequences in silico by mapping overlaps between reads. Bioinformatics then aims to:
- Compare novel sequences to reference genomes and identify variants
- Annotate functional elements like genes, regulatory regions and structural motifs
- Elucidate molecular mechanisms through computational simulations
- In clinical genomics, interpret pathogenic mutations and pharmacogenomics
Deep learning models boost analysis accuracy by discovering subtle patterns difficult for human experts to detect unaided.
Interpretation unlocks insights into evolution, human disease risk, drug response traits, forensic identification and more.
Without converting digital sequences into biological insights, they would lack scientific value.
Can You Arrange the Key Steps Correctly?
To summarize the major DNA technology workflow:
- Extraction Isolate DNA from cells
- PCR Amplification Exponentially multiply DNA copies
- Sequencing Determine the order of As, Ts, Cs and Gs
- Data Analysis Interpret raw sequence data biologically
Can you rearrange these terms into the standard sequence? Let me know if any aspect requires more explanation to comprehend fully.
Interacting helps improve my communication skills for conveying complex topics accessible.
FAQ
Q. What are the terms related to DNA?
A. Nucleic acid.
Q. What is DNA in technical terms?
A. Deoxyribonucleic acid.
Q. What are the technologies involving DNA?
A. Using enzymes to cut and paste pieces of DNA together (recombinant DNA).
Q. What are the examples of DNA technology?
A. DNA sequencing, polymerase chain reaction, DNA cloning, and gel electrophoresis.
Q. What are the 4 words of DNA?
A. Adenine (A), cytosine (C), guanine (G), and thymine (T).
Conclusion:
I hope this comprehensive guide to the standard procedures in DNA technology has helped contextualize recent innovations and breakthroughs in genetics.
By understanding how DNA extraction, amplification, sequencing and analytics are applied, you now have a working knowledge of the methodology behind major scientific achievements and applications from ancestry tracing to precision medicine.
Our abilities to analyze the genome through continued refinement of techniques like next gen sequencing will only grow.
Ultimately, advancing DNA technology brings us closer to unlocking humanity’s shared biological code and improving lives through a deeper grasp of our genetic inheritance.
Please let me know if any part of this journey through the key steps in DNA technology needs more explanation.